A Johns Hopkins-led team has developed a computational model revealing how immune cells can be tweaked to control their ability to absorb or reject external particles. By adjusting receptor clustering on cell surfaces, scientists can now potentially reprogram cells to better distinguish between harmful substances and beneficial ones—including therapeutic nanoparticles. This discovery, funded by the National Science Foundation and published in the Proceedings of the National Academy of Sciences (PNAS), could help improve drug delivery systems, immunization methods, and cancer treatments.
The study is “Receptor clustering tunes and sharpens the selectivity of multivalent binding.”
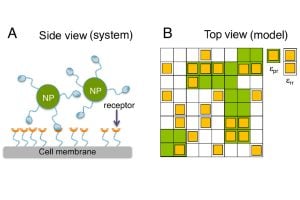
“Typically, cell receptors attach to external agents, such as signaling molecules, viruses or drug-delivery vehicles, and then respond to whatever they’ve attached to, which can lead to miscalculations by the immune system—it can hyper react or under respond, causing illness,” says study leader Tine Curk, an assistant professor of materials science and engineering at the Whiting School of Engineering. “Our findings show that when receptors cluster, they form ‘hot spots’ — places on the cell membrane where particles can bind more securely. This seems to give the cells more control over what they let in or keep out.”
To create their approach, the team used a lattice gas model, a mathematical tool that represents receptors as points on a grid, to study how receptors moved and interacted. Then they ran Monte Carlo simulations, using random sampling to explore possible behaviors and reveal how changes in receptor attraction control whether a cell binds to external molecules.
“We improved a widely used model and applied it to simulations that would reveal if receptors can adjust their responses to each other,” says Curk. “Specifically, we changed interaction energy to make the receptors ‘like’ each other more and measured the effects on the selectivity of binding to external molecules or nanoparticles.”
These simulations confirmed that receptor clustering directly affects how cells bind to these external agents.
“When receptors cluster, they are more likely to bind to external particles. When they don’t cluster, the receptors repel other particles,” says Curk.
Using molecular dynamics simulations, the team then modeled endocytosis—the process by which cells allow particles to enter. They found that when they balanced receptor attraction, the cells selectively absorbed beneficial molecules, like medications, and repelled harmful ones, like viruses.
Curk and the team are now working with Professor of Materials Science and Engineering Kalina Hristova to continue this project with more experimental work.
“Now that we know this is possible, we can trigger clustering in real cells by binding adaptor proteins to the intracellular part of the receptors, through a process called phosphorylation. Understanding receptor clustering could pave the way for enhancements in cancer treatments, drug and vaccine delivery, and more,” Curk said.
The team also includes Zhaoping Xie from the Department of Geriatric Medicine at JHU and collaborators from the United Kingdom and the Chinese Academy of Sciences.